首页 > 代码库 > 中性突变neutral mutation
中性突变neutral mutation
http://baike.baidu.com/view/226162.htm?fr=aladdin
目录
1定义
2历史
3随机漂变假说
4意义
1定义编辑
2历史编辑
3随机漂变假说编辑
4意义编辑
Neutral mutations are changes in DNA sequence that are neither beneficial nor detrimental to the ability of an organism to survive and reproduce. In population genetics, mutationsin which natural selection does not affect the spread of the mutation in a species are termed neutral mutations. Neutral mutations that are inheritable and not linked to any genes under selection will either be lost or will replace all other alleles of the gene. This loss or fixation of the gene proceeds based on random sampling known as genetic drift. A neutral mutation that is in linkage disequilibrium with other alleles that are under selection may proceed to loss or fixation via genetic hitchhiking and/or background selection.
While many mutations in a genome may decrease an organism’s ability to survive and reproduce, also known as fitness, these mutations are selected against and not passed on to future generations. The most commonly observed mutations detectable as variation in the genetic makeup of organisms and populations appear to have no visible effect on the fitness of individuals and are therefore neutral. The identification and study of neutral mutations has led to the development of the neutral theory of molecular evolution. The neutral theory of molecular evolution is an important and often controversial theory proposing that most molecular variation within and among species is essentially neutral and not acted on by selection. Neutral mutations are also the basis for using molecular clocks to identify such evolutionary events as speciation and adaptive or evolutionary radiations.
Contents
[hide]- 1 History
- 2 Impact on evolutionary theory
- 3 Types of neutral mutation
- 3.1 Synonymous mutation of bases
- 3.2 Neutral amino acid substitution
- 4 Identification and measurement of neutrality
- 5 Molecular clocks
- 6 External links
- 7 References
History[edit]
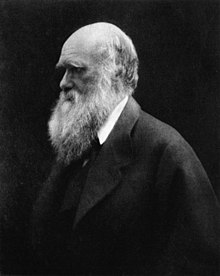
Charles Darwin commented on the idea of neutral mutation in his work, hypothesizing that mutations that do not give an advantage or disadvantage may fluctuate or become fixed apart from natural selection. "Variations neither useful nor injurious would not be affected by natural selection, and would be left either a fluctuating element, as perhaps we see in certain polymorphic species, or would ultimately become fixed, owing to the nature of the organism and the nature of the conditions." While Darwin is widely credited with introducing the idea of natural selection which was the focus of his studies, he also saw the possibility for changes that did not benefit or hurt an organism.[1]
Darwin‘s view of change being mostly driven by traits that provide advantage was widely accepted until the 1960s.[2] While researching mutations that produce nucleotide substitutions in 1968, Motoo Kimura found that the rate of substitution was so high that if each mutation improved fitness, the gap between the most fit and typical genotype would be implausibly large. However, Kimura explained this rapid rate of mutation by suggesting that the majority of mutations were neutral, i.e. had little or no effect on the fitness of the organism. Kimura developed mathematical models of the behavior of neutral mutations subject to random genetic drift in biological populations. This theory has become known as the neutral theory of molecular evolution.[3]
As technology has allowed for better analysis of genomic data, research has continued in this area. While natural selection may encourage adaptation to a changing environment, neutral mutation may push divergence of species due to nearly random genetic drift.[2]
Impact on evolutionary theory[edit]
Neutral mutation has become a part of the neutral theory of molecular evolution, proposed in the 1960s. This theory suggests that neutral mutations are responsible for a large portion of DNA sequence changes in a species. For example, bovine and human insulin, while differing in amino acid sequence are still able to perform the same function. The amino acid substitutions between species were seen therefore to be neutral or not impactful to the function of the protein. Neutral mutation and the neutral theory of molecular evolution are not separate from natural selection but add to Darwin‘s original thoughts. Mutations can give an advantage, create a disadvantage, or make no measurable difference to an organism‘s survival.[4]
A number of observations associated with neutral mutation were predicted in neutral theory including: amino acids with similar biochemical properties should be substituted more often than biochemically different amino acids; synonymous base substitutions should be observed more often than nonsynonymous substitutions; introns should evolve at the same rate as synonymous mutations in coding exons; and pseudogenes should also evolve at a similar rate. These predictions have been confirmed with the introduction of additional genetic data since the theory’s introduction.[2]
Types of neutral mutation[edit]
Synonymous mutation of bases[edit]
When an incorrect nucleotide is inserted during replication or transcription of a coding region, it can affect the eventual translation of the sequence into amino acids. Since multiple codons are used for the same amino acids, a change in a single base may still lead to translation of the same amino acid. This phenomenon is referred to as degeneracy and allows for a variety of codon combinations leading to the same amino acid being produced. For example, the codes TCT, TCC, TCA, TCG, AGT, and AGC all code for the amino acid serine. This can be explained by the wobble concept. Francis Crick proposed this theory to explain why specific tRNA molecules could recognize multiple codons. The area of the tRNA that recognizes the codon called the anticodon is able to bind multiple interchangeable bases at its 5‘ end due to its spacial freedom. A fifth base called inosine can also be substituted on a tRNA and is able to bind with A, U, or C. This flexibility allows for changes in bases in codons leading to translation of the same amino acid.[5] The changing of a base in a codon without the changing of the translated amino acid is called a synonymous mutation. Since the amino acid translated remains the same a synonymous mutation has traditionally been considered a neutral mutation.[6] Some research has suggested that there is bias in selection of base substitution in synonymous mutation. This could be due to selective pressure to improve translation efficiency associated with the most available tRNAs or simply mutational bias.[7] If these mutations influence the rate of translation or an organism’s ability to manufacture protein they may actually influence the fitness of the affected organism.[6]
| nonpolar | polar | basic | acidic | (stop codon) |
| 1st base | 2nd base | 3rd base | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T | C | A | G | ||||||
| T | TTT | (Phe/F)Phenylalanine | TCT | (Ser/S) Serine | TAT | (Tyr/Y)Tyrosine | TGT | (Cys/C)Cysteine | T |
| TTC | TCC | TAC | TGC | C | |||||
| TTA | (Leu/L)Leucine | TCA | TAA | Stop (Ochre) | TGA | Stop (Opal) | A | ||
| TTG | TCG | TAG | Stop (Amber) | TGG | (Trp/W)Tryptophan | G | |||
| C | CTT | CCT | (Pro/P) Proline | CAT | (His/H)Histidine | CGT | (Arg/R)Arginine | T | |
| CTC | CCC | CAC | CGC | C | |||||
| CTA | CCA | CAA | (Gln/Q)Glutamine | CGA | A | ||||
| CTG | CCG | CAG | CGG | G | |||||
| A | ATT | (Ile/I)Isoleucine | ACT | (Thr/T)Threonine | AAT | (Asn/N)Asparagine | AGT | (Ser/S) Serine | T |
| ATC | ACC | AAC | AGC | C | |||||
| ATA | ACA | AAA | (Lys/K)Lysine | AGA | (Arg/R)Arginine | A | |||
| ATG[A] | (Met/M)Methionine | ACG | AAG | AGG | G | ||||
| G | GTT | (Val/V) Valine | GCT | (Ala/A) Alanine | GAT | (Asp/D)Aspartic acid | GGT | (Gly/G)Glycine | T |
| GTC | GCC | GAC | GGC | C | |||||
| GTA | GCA | GAA | (Glu/E)Glutamic acid | GGA | A | ||||
| GTG | GCG | GAG | GGG | G | |||||
- A The codon ATG both codes for methionine and serves as an initiation site: the first ATG in an mRNA‘s coding region is where translation into protein begins.[8]
Neutral amino acid substitution[edit]
While substitution of a base in a noncoding area of a genome may make little difference and be considered neutral, base substitutions in or around genes may impact the organism. Some base substitutions lead to synonymous mutation and no difference in the amino acid translated as noted above. However, a base substitution can also change the genetic code so that a different amino acid is translated. This sort of substitution usually has a negative effect on the protein being formed and will be eliminated from the population throughpurifying selection. However, if the change has a positive influence, the mutation may become more and more common in a population until it becomes a fixed genetic piece of that population. Organisms changing via these two options comprise the classic view of natural selection. A third possibility is that the amino acid substitution makes little or no positive or negative difference to the affected protein.[9] Proteins demonstrate some tolerance to changes in amino acid structure. This is somewhat dependent on where in the protein the substitution takes place. If it occurs in an important structural area or in the active site, one amino acid substitution may inactivate or substantially change the functionality of the protein. Substitutions in other areas may be nearly neutral and drift randomly over time.[10]
Identification and measurement of neutrality[edit]
Neutral mutations are measured in population and evolutionary genetics often by looking at variation in populations. These have been measured historically by gel electrophoresis to determine allozyme frequencies.[11] Statistical analyses of this data is used to compare variation to predicted values based on population size, mutation rates and effective population size. Early observations that indicated higher than expected heterozygosity and overall variation within the protein isoforms studied, drove arguments as to the role of selection in maintaining this variation versus the existence of variation through the effects of neutral mutations arising and their random distribution due to genetic drift.[12][13][14] The accumulation of data based on observed polymorphism led to the formation of the neutral theory of evolution.[12] According to the neutral theory of evolution, the rate of fixation in a population of a neutral mutation will be directly related to the rate of formation of the neutral allele.[15]
In Kimura’s original calculations, mutations with |2 Ns|<1 or |s|≤1/(2N) are defined as neutral.[12][14] In this equation, N is the effective population size and is a quantitative measurement of the ideal population size that assumes such constants as equal sex ratios and no emigration, migration, mutation nor selection.[16] Conservatively, it is often assumed that effective population size is approximately one fifth of the total population size.[17] s is the selection coefficient and is a value between 0 and 1. It is a measurement of the contribution of a genotype to the next generation where a value of 1 would be completely selected against and make no contribution and 0 is not selected against at all.[18] This definition of neutral mutation has been criticized due to the fact that very large effective population sizes can make mutations with little selection coefficients appear non neutral. Additionally, mutations with high selection coefficients can appear neutral in very small populations.[14] The testable hypothesis of Kimura and others showed that polymorphism within species are approximately that which would be expected in a neutral evolutionary model.[14][19][20]
For many molecular biology approaches, as opposed to mathematical genetics, neutral mutations are generally assumed to be those mutations which cause no appreciable effect on gene function. This simplification eliminates the effect of minor allelic differences in fitness and avoids problems when selection has only a minor effect.[14]
Early convincing evidence of this definition of neutral mutation was shown through the lower mutational rates in functionally important parts of genes such as cytochrome c versus less important parts[21] and the functionally interchangeable nature of mammalian cytochrome c in in vitro studies.[22] Nonfunctional pseudogenes provide more evidence for the role of neutral mutations in evolution. The rates of mutation in mammalian globin pseudogenes has been shown to be much higher than rates in functional genes.[23][24] According to neo-Darwinian evolution, such mutations should rarely exist as these sequences are functionless and positive selection would not be able to operate.[14]
The McDonald-Kreitman test[25] has been used to study selection over long periods of evolutionary time. This is a statistical test that compares polymorphism in neutral and functional sites and estimates what fraction of substitutions have been acted on by positive selection.[26] The test often uses synonymous substitutions in protein coding genes as the neutral component, however, synonymous mutations have been shown to be under purifying selection in many instances.[27][28]
Molecular clocks[edit]
Molecular clocks can be used to estimate the amount of time since divergence of two species and for placing evolutionary events in time.[29] Pauling and Zuckerkandl, proposed the idea of the molecular clock in 1962 based on the observation that the random mutation process occurs at an approximate constant rate. Individual proteins were shown to have linear rates of amino acid changes over evolutionary time.[30] Despite controversy from some biologists arguing that morphological evolution would not proceed at a constant rate, many amino acid changes were shown to accumulate in a constant fashion. Kimura and Ohta explained these rates as part of the framework of the neutral theory. These mutations were reasoned to be neutral as positive selection should be rare and deleterious mutations should be eliminated quickly from a population.[31] By this reasoning, the accumulation of these neutral mutations should only be influenced by the mutation rate. Therefore, the neutral mutation rate in individual organisms should match the molecular evolution rate in species over evolutionary time. The neutral mutation rate is affected by the amount of neutral sites in a protein or DNA sequence versus the amount of mutation in sites that are functionally constrained. By quantifying these neutral mutations in protein and/or DNA and comparing them between species or other groups of interest, rates of divergence can be determined.[29][32]
Molecular clocks have caused controversy as to their dates derived on events such as explosive radiations seen after extinction events like the Cambrian explosion and the radiations of mammals and birds. Two fold differences exist in dates from molecular clocks and the fossil record. While some paleontologists argue that molecular clocks are systemically inaccurate, others attribute the discrepancies to lack of robust fossil data and bias in sampling.[33] While not without constancy and discrepancies with the fossil record, the data from molecular clocks has shown how evolution is dominated by the mechanisms of a neutral model and is less influenced by the action of natural selection.
中性突变neutral mutation
